Databases¶
Modules¶
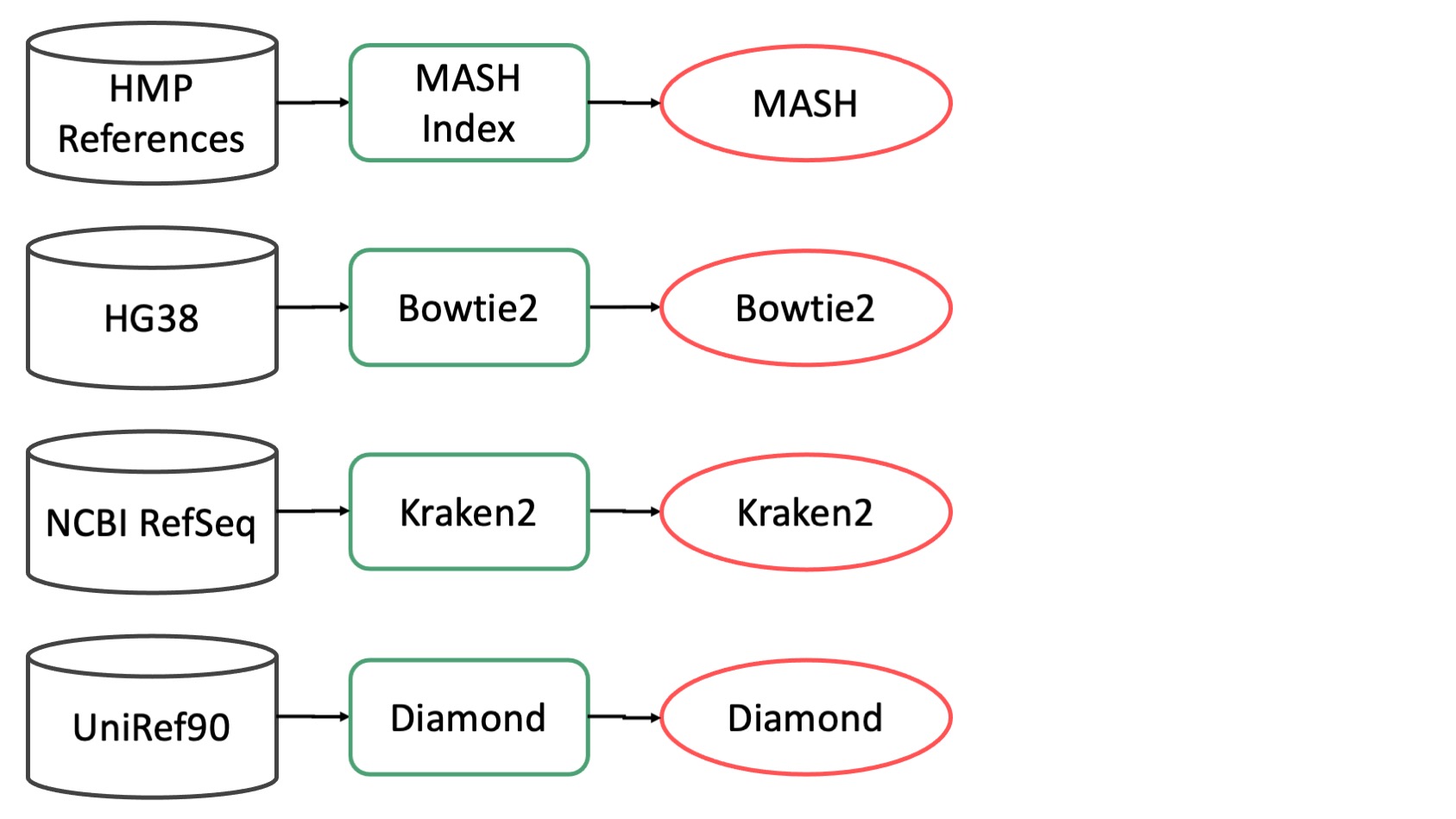
Mouse Removal Database¶
The CAP uses mm39 for mouse removal. The raw (fasta) genome is indexed using Bowtie2.
Human Removal Database¶
The CAP uses Hg38 with alternate contigs. for human removal. The raw (fasta) genome is indexed using Bowtie2. Th rationale for picking this genome is based on this.